Data Cleaning

One of the more common tasks of a data scientists is that of data cleaning. Unfortunately, not all datasets come to us ready for analysis out of the box. Some require a little work to be ready, and some require a little more. I recently came across a dataset that was REALLY in need of some serious cleaning. It is a dataset (found here) that catalogued academic studies and their cost to publish them openly. The fields include one or two ID numbers corresponding to each paper, the paper title, journal, publisher, and total cost in GBP of making the study open. Below I’ll go through and annotate my steps to clean (partially) this dataset. I say partially because there are so many unique cases of entries that need cleaning in the Journal and Publisher columns that to really clean requires more work that can be encapsulated in a blog post.
Note: you might argue that I could have done this work in fewer steps/lines of code. Sure, you’re right! I make no claims that this is the most terse or pythonic solution to cleaning the dataset. This post reflects my work in cleaning the dataset as I was doing the work for the first time. Of course, ex post facto, it’s much easier to think of a way to do the same task better.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
df = pd.read_excel("University returns_for_figshare_FINAL.xlsx")
df.head()
| PMID/PMCID | Publisher | Journal title | Article title | COST (£) charged to Wellcome (inc VAT when charged) | |
|---|---|---|---|---|---|
| 0 | PMC3378987\n | Elsevier | Academy of Nutrition and Dietetics | Parent support and parent mediated behaviours ... | 2379.54 |
| 1 | PMCID: PMC3780468 | ACS (Amercian Chemical Society) Publications | ACS Chemical Biology | A Novel Allosteric Inhibitor of the Uridine Di... | 1294.59 |
| 2 | PMCID: PMC3621575 | ACS (Amercian Chemical Society) Publications | ACS Chemical Biology | Chemical proteomic analysis reveals the drugab... | 1294.78 |
| 3 | NaN | American Chemical Society | ACS Chemical Biology | Discovery of β2 Adrenergic Receptor Ligands Us... | 947.07 |
| 4 | PMID: 24015914 PMC3833349 | American Chemical Society | ACS Chemical Biology | Discovery of an allosteric inhibitor binding s... | 1267.76 |
df.columns=["PMCID","Publisher","Journal Title","Article Title","TotalCost"]
df.head()
| PMCID | Publisher | Journal Title | Article Title | TotalCost | |
|---|---|---|---|---|---|
| 0 | PMC3378987\n | Elsevier | Academy of Nutrition and Dietetics | Parent support and parent mediated behaviours ... | 2379.54 |
| 1 | PMCID: PMC3780468 | ACS (Amercian Chemical Society) Publications | ACS Chemical Biology | A Novel Allosteric Inhibitor of the Uridine Di... | 1294.59 |
| 2 | PMCID: PMC3621575 | ACS (Amercian Chemical Society) Publications | ACS Chemical Biology | Chemical proteomic analysis reveals the drugab... | 1294.78 |
| 3 | NaN | American Chemical Society | ACS Chemical Biology | Discovery of β2 Adrenergic Receptor Ligands Us... | 947.07 |
| 4 | PMID: 24015914 PMC3833349 | American Chemical Society | ACS Chemical Biology | Discovery of an allosteric inhibitor binding s... | 1267.76 |
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 2127 entries, 0 to 2126
Data columns (total 5 columns):
PMCID 1928 non-null object
Publisher 2127 non-null object
Journal Title 2126 non-null object
Article Title 2127 non-null object
TotalCost 2127 non-null float64
dtypes: float64(1), object(4)
memory usage: 83.2+ KB
Let’s start fixing the PMCID column. The first things I notice just from the head() are newlines actually in the value, “PMCID: “ still there when the column header says what the value is, and null values.
df["PMCID"].fillna("Unknown",inplace=True)
df["PMCID"]
0 PMC3378987\n
1 PMCID: PMC3780468
2 PMCID: PMC3621575
3 Unknown
4 PMID: 24015914 PMC3833349
5 : PMC3805332
6 PMCID:\n PMC3656742\n
7 PMCID: 3584654
8 23373658
9 PMCID:\n PMC3727331\n
10 PMCID: PMC3565438
11 PMCID: PMC3668577
12 PMCID: PMC3606566
13 \n PMC3498934
14 PMID:22993091 PMC3447403
15 PMC3087623
16 PMC3808818
17 PMID: 23828613 (July 2013 Epub)
18 PMC3374517
19 PMC3549237
20 Unknown
21 PMC3661931
22 3535376
23 PMC3798121
24 21624095 PMCID: PMC3734623
25 In Process
26 23734913
27 Unknown
28 Unknown
29 Unknown
...
2097 PMCID:\n PMC3508281\n\n
2098 PMC3717178
2099 PMC3775257
2100 PMC3558801
2101 Unknown
2102 PMC3770928
2103 PMC3627817
2104 PMCID:\n PMC3759846
2105 PMCID:\n PMC3608034
2106 Unknown
2107 PMCID: PMC3599165
2108 PMC3763375
2109 PMID: 24035434
2110 PMC3763374
2111 PMC3404461
2112 PMCID: PMC3740234
2113 23117109
2114 PMC3757156
2115 Unknown
2116 PMC3611597
2117 PMC3786614
2118 PMC3716626
2119 PMCID: PMC3791421
2120 23562481
2121 PMC3190389
2122 23201205
2123 pub Aug 2013
2124 23200744 PMC3552157
2125 PMC3472342\n\n
2126 PMCID: PMC3600532
Name: PMCID, Length: 2127, dtype: object
OK, it seems from looking at the pubmed website, there are two identifiers, PMID and PMCID. The former is an 8-digit number and the latter is a 7-digit number preceded by “PMC”. The entries in this column that are just 8-digit numbers I’ll assume are PMIDs. I propose to just make this column into two separate columns: PMID and PMCID
def extractNonPMCID(row):
id = str(row.PMCID)
if len(id) < 4:
return id
elif id[0:3] == "PMC":
return "Unknown"
else:
return id
def fixPMCID(id):
id = str(id)
if len(id) < 4:
return "Unknown"
elif id[0:3] == "PMC":
return id
else:
return "Unknown"
df["PMID"] = df.apply(extractNonPMCID,axis=1)
df["PMCID"] = df["PMCID"].apply(fixPMCID)
df["PMCID"]
0 PMC3378987\n
1 PMCID: PMC3780468
2 PMCID: PMC3621575
3 Unknown
4 Unknown
5 Unknown
6 PMCID:\n PMC3656742\n
7 PMCID: 3584654
8 Unknown
9 PMCID:\n PMC3727331\n
10 PMCID: PMC3565438
11 PMCID: PMC3668577
12 PMCID: PMC3606566
13 Unknown
14 Unknown
15 PMC3087623
16 PMC3808818
17 Unknown
18 PMC3374517
19 PMC3549237
20 Unknown
21 PMC3661931
22 Unknown
23 PMC3798121
24 Unknown
25 Unknown
26 Unknown
27 Unknown
28 Unknown
29 Unknown
...
2097 PMCID:\n PMC3508281\n\n
2098 PMC3717178
2099 PMC3775257
2100 PMC3558801
2101 Unknown
2102 PMC3770928
2103 PMC3627817
2104 PMCID:\n PMC3759846
2105 PMCID:\n PMC3608034
2106 Unknown
2107 PMCID: PMC3599165
2108 PMC3763375
2109 Unknown
2110 PMC3763374
2111 PMC3404461
2112 PMCID: PMC3740234
2113 Unknown
2114 PMC3757156
2115 Unknown
2116 PMC3611597
2117 PMC3786614
2118 PMC3716626
2119 PMCID: PMC3791421
2120 Unknown
2121 PMC3190389
2122 Unknown
2123 Unknown
2124 Unknown
2125 PMC3472342\n\n
2126 PMCID: PMC3600532
Name: PMCID, Length: 2127, dtype: object
df["PMID"]
0 Unknown
1 Unknown
2 Unknown
3 Unknown
4 PMID: 24015914 PMC3833349
5 : PMC3805332
6 Unknown
7 Unknown
8 23373658
9 Unknown
10 Unknown
11 Unknown
12 Unknown
13 \n PMC3498934
14 PMID:22993091 PMC3447403
15 Unknown
16 Unknown
17 PMID: 23828613 (July 2013 Epub)
18 Unknown
19 Unknown
20 Unknown
21 Unknown
22 3535376
23 Unknown
24 21624095 PMCID: PMC3734623
25 In Process
26 23734913
27 Unknown
28 Unknown
29 Unknown
...
2097 Unknown
2098 Unknown
2099 Unknown
2100 Unknown
2101 Unknown
2102 Unknown
2103 Unknown
2104 Unknown
2105 Unknown
2106 Unknown
2107 Unknown
2108 Unknown
2109 PMID: 24035434
2110 Unknown
2111 Unknown
2112 Unknown
2113 23117109
2114 Unknown
2115 Unknown
2116 Unknown
2117 Unknown
2118 Unknown
2119 Unknown
2120 23562481
2121 Unknown
2122 23201205
2123 pub Aug 2013
2124 23200744 PMC3552157
2125 Unknown
2126 Unknown
Name: PMID, Length: 2127, dtype: object
Wow, this really needed cleaning all right! Let’s first get the PMCID column in order, then we’ll attend to the PMID column (perhaps replacing some PMCID values it seems).
def purgeColons(id):
s = id.split()
if len(s) < 2:
return id
else:
if s[1][0:3] == "PMC":
return s[1]
else:
return "PMC"+s[1]
df["PMCID"] = df["PMCID"].apply(purgeColons)
# I saw a couple entries with two newlines and with a apsce
df["PMCID"] = df["PMCID"].apply( lambda x : x.strip("\n").strip("\n").strip(" ") )
df["PMCID"]
0 PMC3378987
1 PMC3780468
2 PMC3621575
3 Unknown
4 Unknown
5 Unknown
6 PMC3656742
7 PMC3584654
8 Unknown
9 PMC3727331
10 PMC3565438
11 PMC3668577
12 PMC3606566
13 Unknown
14 Unknown
15 PMC3087623
16 PMC3808818
17 Unknown
18 PMC3374517
19 PMC3549237
20 Unknown
21 PMC3661931
22 Unknown
23 PMC3798121
24 Unknown
25 Unknown
26 Unknown
27 Unknown
28 Unknown
29 Unknown
...
2097 PMC3508281
2098 PMC3717178
2099 PMC3775257
2100 PMC3558801
2101 Unknown
2102 PMC3770928
2103 PMC3627817
2104 PMC3759846
2105 PMC3608034
2106 Unknown
2107 PMC3599165
2108 PMC3763375
2109 Unknown
2110 PMC3763374
2111 PMC3404461
2112 PMC3740234
2113 Unknown
2114 PMC3757156
2115 Unknown
2116 PMC3611597
2117 PMC3786614
2118 PMC3716626
2119 PMC3791421
2120 Unknown
2121 PMC3190389
2122 Unknown
2123 Unknown
2124 Unknown
2125 PMC3472342
2126 PMC3600532
Name: PMCID, Length: 2127, dtype: object
OK let’s see what remains to be corrected in the PMCID column
df.PMCID[(df.PMCID != "Unknown") & ~(df.PMCID.str.startswith("PMC3"))]
111 PMC:3697379
158 PMCID:PMC3542821
235 PMC2843621
357 PMCID:PMC3718323
384 PMC
392 PMCID:PMC3778892
486 PMC
613 PMC(Available
739 PMC2881129
831 PMCin
880 PMCID:3106451
901 PMCID:PMC3268392
963 PMC2757965
1037 PMCID:PMC3708113
1077 PMCID:PMC3778837
1143 PMCID:PMC3759848
1178 PMCID:PMC3728563
1361 PMCID:PMC3709497
1371 PMCID:PMC3572772
1397 PMC2877799
1404 PMCID:
1464 PMC2779337
1533 PMCin
1570 PMCID:PMC3715547)
1638 PMCID:PMC3706442
1639 PMCID:PMC3711905
1778 PMCis
2008 PMCID:PMC3549627
2022 PMCID3135804
2031 PMC2955965
2038 PMCID:PMC3573229
2039 PMCID:PMC3755220
2064 PMCID:PMC3677413
2078 PMCID:PMC3731785
Name: PMCID, dtype: object
def corrPMCIDEdge(id):
id = id.replace("PMCID:","")
id = id.replace("PMCID","PMC")
id = id.replace("PMC:","")
id = id.replace("PMCin","Unknown")
id = id.replace("PMCis","Unknown")
if id == "PMC":
return "Unknown"
elif id == "PMC(Available":
return "Unknown"
else:
return id
df["PMCID"]=df["PMCID"].apply(corrPMCIDEdge)
df.PMCID[(df.PMCID != "Unknown") & ~(df.PMCID.str.startswith("PMC3"))]
111 3697379
235 PMC2843621
739 PMC2881129
880 3106451
963 PMC2757965
1397 PMC2877799
1404
1464 PMC2779337
2031 PMC2955965
Name: PMCID, dtype: object
# Do the rest by hand
df.at[111,"PMCID"] = "PMC3697379"
df.at[880,"PMCID"] = "PMC3106451"
df.at[1404,"PMCID"] = "Unknown"
df["PMCID"] = [str(x) for x in df.PMCID]
df[~(df.PMCID.str.startswith("PMC3")) & (df.PMCID != "Unknown")]["PMCID"]
235 PMC2843621
739 PMC2881129
963 PMC2757965
1397 PMC2877799
1464 PMC2779337
2031 PMC2955965
Name: PMCID, dtype: object
OK PMCID is clean! Let’s now tackle PMID
df.PMID
0 Unknown
1 Unknown
2 Unknown
3 Unknown
4 PMID: 24015914 PMC3833349
5 : PMC3805332
6 Unknown
7 Unknown
8 23373658
9 Unknown
10 Unknown
11 Unknown
12 Unknown
13 \n PMC3498934
14 PMID:22993091 PMC3447403
15 Unknown
16 Unknown
17 PMID: 23828613 (July 2013 Epub)
18 Unknown
19 Unknown
20 Unknown
21 Unknown
22 3535376
23 Unknown
24 21624095 PMCID: PMC3734623
25 In Process
26 23734913
27 Unknown
28 Unknown
29 Unknown
...
2097 Unknown
2098 Unknown
2099 Unknown
2100 Unknown
2101 Unknown
2102 Unknown
2103 Unknown
2104 Unknown
2105 Unknown
2106 Unknown
2107 Unknown
2108 Unknown
2109 PMID: 24035434
2110 Unknown
2111 Unknown
2112 Unknown
2113 23117109
2114 Unknown
2115 Unknown
2116 Unknown
2117 Unknown
2118 Unknown
2119 Unknown
2120 23562481
2121 Unknown
2122 23201205
2123 pub Aug 2013
2124 23200744 PMC3552157
2125 Unknown
2126 Unknown
Name: PMID, Length: 2127, dtype: object
df.PMID = [str(x) for x in df.PMID]
df.PMID[df.PMID.str.contains("PMC")]
4 PMID: 24015914 PMC3833349
5 : PMC3805332
13 \n PMC3498934
14 PMID:22993091 PMC3447403
24 21624095 PMCID: PMC3734623
31 21948184 PMC3528370
33 PMID: 23672932 PMC3684112
54 22971149 PMC3466778
64 PMID: 23907068 PMC3837358
73 PMID: 22898127 PMC3830178
74 PMID: 23159264 PMC3834139
90 23670821 PMC3738939
93 PMID:22961729 PMC3547296
94 PMID:22730171 PMC3556687
95 PMID:21472932 PMC3555362
107 PMID:23089748 PMC3535899
108 PMID: 23856774 PMC3811480
110 PMID:23817378 PMCID: PMC3754341
123 23740368 PMC3840700
124 23460124 PMC3715109
132 22302008 PMC3405838
137 23025831 PMC3462035
138 23731076 PMC3670620
139 23244291 PMC3569044
147 PMID:22326920 PMC3480643
149 PMID: 20377523 PMC3685215
150 PMID: 23009366 PMC3685217
162 PMID: 23356304 PMC3561678
163 PMID: 23697937 PMC3685327
164 PMCID:\n PMC3554041\n
...
1919 PMID: 23407782 PMC3676742
1920 PMID: 23299096 PMC3636441
1921 PMID: 23681165 PMC3824307
1923 PMID: 23332733 PMC3712184
1935 22777780 PMCID: PMC3568905
1950 23422316 PMCID: PMC3710354
1955 PMID: 22704639 PMC3485564
1956 PMID: 21764562 PMC3485562
1957 23230506 PMC3516806
1969 23698510 PMC3812900
1993 PMID23287458 PMC3605587
1996 23356787 PMC3813989
1999 22618994 PMC344470
2010 PMID:23623732 PMC3791409
2020 23375655 PMC3567274
2025 PMID: 23282150 PMC3610541
2030 PMID: 24076655 PMC3817463
2033 PMID: 23371065 /PMCID: PMC3633812
2046 23698002 PMC3696650
2049 PMID: 23125445 PMC3532835
2051 23576705, PMC3727040
2052 23804098 PMCID: PMC3724994
2053 23825402 PMC3528370
2062 PMID: 23041239 /PMCID: PMC3490334
2067 PMID: 23709760 /PMCID: PMC3756442
2070 22901061 PMC3533787
2074 PMID: 19948006 PMC3551259
2080 PMID: 21940062 PMC3814186
2083 PMID:23265842 PMC3569712
2124 23200744 PMC3552157
Name: PMID, Length: 276, dtype: object
pmcs = df.PMID[df.PMID.str.contains("PMC")].apply(lambda x : x.split()[-1])
pmcs[~pmcs.str.startswith("PMC3")]
632 PMC
633 PMC
784 (PMCID:PMC3757918)
1068 PMCID3274377
1273 -PMC3839404
1456 -PMC3661980
1537 3679597
1591 24039607/PMC3764205
1687 24124519/PMC3790821
Name: PMID, dtype: object
pmcs.drop(632,axis=0,inplace=True)
pmcs.drop(633,axis=0,inplace=True)
pmcs.at[784]="PMC3757918"
pmcs.at[1068]="PMC3274377"
pmcs.at[1273]="PMC3839404"
pmcs.at[1456]="PMC3661980"
pmcs.at[1537]="PMC3679597"
pmcs.at[1591]="PMC3764205"
pmcs.at[1687]="PMC3790821"
# pmcs.index
for i in pmcs.index:
print df.PMCID[i],pmcs[i]
Unknown PMC3833349
Unknown PMC3805332
Unknown PMC3498934
Unknown PMC3447403
Unknown PMC3734623
Unknown PMC3528370
Unknown PMC3684112
Unknown PMC3466778
Unknown PMC3837358
Unknown PMC3830178
Unknown PMC3834139
Unknown PMC3738939
Unknown PMC3547296
Unknown PMC3556687
Unknown PMC3555362
Unknown PMC3535899
Unknown PMC3811480
Unknown PMC3754341
Unknown PMC3840700
Unknown PMC3715109
Unknown PMC3405838
Unknown PMC3462035
Unknown PMC3670620
Unknown PMC3569044
Unknown PMC3480643
Unknown PMC3685215
Unknown PMC3685217
Unknown PMC3561678
Unknown PMC3685327
Unknown PMC3554041
Unknown PMC3413243
Unknown PMC3714738
Unknown PMC3630740
Unknown PMC3694669
Unknown PMC3381227
Unknown PMC3736666
Unknown PMC3814418
Unknown PMC3790949
Unknown PMC3653566
Unknown PMC3790942
Unknown PMC3846689
Unknown PMC3612817
Unknown PMC3525062
Unknown PMC3573228
Unknown PMC3596649
Unknown PMC3687655
Unknown PMC3724117
Unknown PMC3730114
Unknown PMC3491872
Unknown PMC3674087
Unknown PMC3619236
Unknown PMC3601332
Unknown PMC3510442
Unknown PMC3689915
Unknown PMC3753470
Unknown PMC3599488
Unknown PMC3599488
Unknown PMC3715701
Unknown PMC3539454
Unknown PMC3625746
Unknown PMC3792637
Unknown PMC3401426
Unknown PMC3569615
Unknown PMC3743159
Unknown PMC3807794
Unknown PMC3566545
Unknown PMC3629559
Unknown PMC3706957
Unknown PMC3502867
Unknown PMC3485558
Unknown PMC3593001
Unknown PMC3521961
Unknown PMC3709123
Unknown PMC3381227
Unknown PMC3778978
Unknown PMC3849851
Unknown PMC3638368
Unknown PMC3427858
Unknown PMC3629561
Unknown PMC3744751
Unknown PMC3630358
Unknown PMC3791366
Unknown PMC3701237
Unknown PMC3707360
Unknown PMC3669511
Unknown PMC3547489
Unknown PMC3657127
Unknown PMC3781703
Unknown PMC3744815
Unknown PMC3784797
Unknown PMC3745826
Unknown PMC3630327
Unknown PMC3659828
Unknown PMC3572582
Unknown PMC3564407
Unknown PMC3734648
Unknown PMC3806363
Unknown PMC3642154
Unknown PMC3668512
Unknown PMC3654214
Unknown PMC3693486
Unknown PMC3696884
Unknown PMC3664024
Unknown PMC3638342
Unknown PMC3487588
Unknown PMC3591252
Unknown PMC3727344
Unknown PMC3744754
Unknown PMC3757918
Unknown PMC3652415
Unknown PMC3842180
Unknown PMC3710569
Unknown PMC3824577
Unknown PMC3497410
Unknown PMC3536122
Unknown PMC3631001
Unknown PMC3819985
Unknown PMC3566929
Unknown PMC3821636
Unknown PMC3734734
Unknown PMC3504370
Unknown PMC3634610
Unknown PMC3730248
Unknown PMC3672848
Unknown PMC3506128
Unknown PMC3646402
Unknown PMC3601604
Unknown PMC3797641
Unknown PMC3561570
Unknown PMC3493908
Unknown PMC3543027
Unknown PMC3585060
Unknown PMC3711325
Unknown PMC3531760
Unknown PMC3516716
Unknown PMC3591641
Unknown PMC3668706
Unknown PMC3750171
Unknown PMC3576117
Unknown PMC3662417
Unknown PMC3746124
Unknown PMC3828593
Unknown PMC3562487
Unknown PMC3651585
Unknown PMC3784648
Unknown PMC3813945
Unknown PMC3708032
Unknown PMC3851688
Unknown PMC3756433
Unknown PMC3576129
Unknown PMC3749055
Unknown PMC3709586
Unknown PMC3709636
Unknown PMC3709573
Unknown PMC3709587
Unknown PMC3542733
Unknown PMC3709688
Unknown PMC3607399
Unknown PMC3619528
Unknown PMC3540259
Unknown PMC3654748
Unknown PMC3274377
Unknown PMC3513931
Unknown PMC3798117
Unknown PMC3579312
Unknown PMC3709824
Unknown PMC3593212
Unknown PMC3535724
Unknown PMC3664183
Unknown PMC3544867
Unknown PMC3664272
Unknown PMC3601669
Unknown PMC3593678
Unknown PMC3586169
Unknown PMC3754066
Unknown PMC3554137
Unknown PMC3457148
Unknown PMC3807398
Unknown PMC3807390
Unknown PMC3641668
Unknown PMC3677138
Unknown PMC3691584
Unknown PMC3440593
Unknown PMC3520108
Unknown PMC3839404
Unknown PMC3836489
Unknown PMC3495574
Unknown PMC3581773
Unknown PMC3820028
Unknown PMC3625108
Unknown PMC3587388
Unknown PMC3529057
Unknown PMC3528594
Unknown PMC3826647
Unknown PMC3514496
Unknown PMC3685308
Unknown PMC3782194
Unknown PMC3532594
Unknown PMC3757163
Unknown PMC3707011
Unknown PMC3480648
Unknown PMC3555187
Unknown PMC3580290
Unknown PMC3542422
Unknown PMC3611599
Unknown PMC3661980
Unknown PMC3675483
Unknown PMC3753647
Unknown PMC3401426
Unknown PMC3643581
Unknown PMC3834809
Unknown PMC3245117
Unknown PMC3492912
Unknown PMC3798095
Unknown PMC3717798
Unknown PMC3809720
Unknown PMC3679597
Unknown PMC3764205
Unknown PMC3542187
Unknown PMC3541403
Unknown PMC3558488
Unknown PMC3487758
Unknown PMC3790821
Unknown PMC3733773
Unknown PMC3716603
Unknown PMC3689675
Unknown PMC3556037
Unknown PMC3491040
Unknown PMC3519623
Unknown PMC3632543
Unknown PMC3686795
Unknown PMC3762823
Unknown PMC3576415
Unknown PMC3585132
Unknown PMC3596319
Unknown PMC3841754
Unknown PMC3712186
Unknown PMC3824070
Unknown PMC3465389
Unknown PMC3529034
Unknown PMC3562850
Unknown PMC3568321
Unknown PMC3581986
Unknown PMC3569869
Unknown PMC3676742
Unknown PMC3636441
Unknown PMC3824307
Unknown PMC3712184
Unknown PMC3568905
Unknown PMC3710354
Unknown PMC3485564
Unknown PMC3485562
Unknown PMC3516806
Unknown PMC3812900
Unknown PMC3605587
Unknown PMC3813989
Unknown PMC344470
Unknown PMC3791409
Unknown PMC3567274
Unknown PMC3610541
Unknown PMC3817463
Unknown PMC3633812
Unknown PMC3696650
Unknown PMC3532835
Unknown PMC3727040
Unknown PMC3724994
Unknown PMC3528370
Unknown PMC3490334
Unknown PMC3756442
Unknown PMC3533787
Unknown PMC3551259
Unknown PMC3814186
Unknown PMC3569712
Unknown PMC3552157
for i in pmcs.index:
df.at[i,"PMCID"]=pmcs[i]
We were able to find PMCIDs for lots of the Unknowns using the PMID entries!
df.PMID[df.PMID != "Unknown"]
4 PMID: 24015914 PMC3833349
5 : PMC3805332
8 23373658
13 \n PMC3498934
14 PMID:22993091 PMC3447403
17 PMID: 23828613 (July 2013 Epub)
22 3535376
24 21624095 PMCID: PMC3734623
25 In Process
26 23734913
31 21948184 PMC3528370
32 23340916
33 PMID: 23672932 PMC3684112
35 print published August 2013
37 3748854
39 3633780
40 In Process
51 23514390
52 3516067
53 23599091
54 22971149 PMC3466778
61 3813311
64 PMID: 23907068 PMC3837358
69 3567269
71 published Sept 2013
72 3947303
73 PMID: 22898127 PMC3830178
74 PMID: 23159264 PMC3834139
75 3708126
77 PMID: 23409903 23409903
...
2018 PMID: 23649934
2020 23375655 PMC3567274
2025 PMID: 23282150 PMC3610541
2030 PMID: 24076655 PMC3817463
2033 PMID: 23371065 /PMCID: PMC3633812
2036 23253866
2037 Not yet available
2046 23698002 PMC3696650
2049 PMID: 23125445 PMC3532835
2051 23576705, PMC3727040
2052 23804098 PMCID: PMC3724994
2053 23825402 PMC3528370
2062 PMID: 23041239 /PMCID: PMC3490334
2063 PMID : 23773811
2067 PMID: 23709760 /PMCID: PMC3756442
2070 22901061 PMC3533787
2073 3744763
2074 PMID: 19948006 PMC3551259
2075 PMID: 23305527
2079 pub Aug 2013
2080 PMID: 21940062 PMC3814186
2083 PMID:23265842 PMC3569712
2086 print in press
2096 22364555
2109 PMID: 24035434
2113 23117109
2120 23562481
2122 23201205
2123 pub Aug 2013
2124 23200744 PMC3552157
Name: PMID, Length: 741, dtype: object
Whoa, there are lots of different cases to consider…Looks like one of the most common is
PMID: XXXXXXXX PMCYYYYYYY
Let’s tackle those
df.PMID[df.PMID.str.contains("PMC")] = df.PMID[df.PMID.str.contains("PMC")].apply(lambda x : x.replace("/","")) #drop slashes
def case1(id):
s = id.split()
if len(s) != 3:
return id
elif s[0] == "PMID:" and s[2][0:3]=="PMC":
# print s[1]
return s[1]
else:
return id
df["PMID"]=df.PMID.apply(case1)
/Users/skaplan/Library/Python/2.7/lib/python/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
"""Entry point for launching an IPython kernel.
Now let’s deal with the case
XXXXXXXX PMCYYYYYYY
df.PMID[df.PMID.str.contains("PMC")]
5 : PMC3805332
13 \n PMC3498934
14 PMID:22993091 PMC3447403
24 21624095 PMCID: PMC3734623
31 21948184 PMC3528370
54 22971149 PMC3466778
90 23670821 PMC3738939
93 PMID:22961729 PMC3547296
94 PMID:22730171 PMC3556687
95 PMID:21472932 PMC3555362
107 PMID:23089748 PMC3535899
110 PMID:23817378 PMCID: PMC3754341
123 23740368 PMC3840700
124 23460124 PMC3715109
132 22302008 PMC3405838
137 23025831 PMC3462035
138 23731076 PMC3670620
139 23244291 PMC3569044
147 PMID:22326920 PMC3480643
164 PMCID:\n PMC3554041\n
165 PMC3413243
171 23458425 PMC3630740
205 22738332 PMC3381227
211 23931322 PMCID: PMC3736666
212 23877967 PMC3814418
227 23002116 PMC3790949
229 23255554 PMC3653566
230 23043070 PMC3790942
321 23246923 PMC3573228
338 23121507 PMCID: PMC3596649
...
1785 PMID: 24023766 PMCID: PMC3762823
1793 \n PMC3576415
1825 PMID: 23468629 PMCID: PMC3585132
1841 \nPMC3596319
1854 23924687 PMC3841754
1856 PMID: 23375993 PMCID: PMC3712186
1863 PMCID: PMC3465389
1880 23213245 PMC3529034
1885 23341602 PMC3568321
1892 PMID:23391734 PMC3581986
1907 PMID:22997008 PMC3569869
1935 22777780 PMCID: PMC3568905
1950 23422316 PMCID: PMC3710354
1957 23230506 PMC3516806
1969 23698510 PMC3812900
1993 PMID23287458 PMC3605587
1996 23356787 PMC3813989
1999 22618994 PMC344470
2010 PMID:23623732 PMC3791409
2020 23375655 PMC3567274
2033 PMID: 23371065 PMCID: PMC3633812
2046 23698002 PMC3696650
2051 23576705, PMC3727040
2052 23804098 PMCID: PMC3724994
2053 23825402 PMC3528370
2062 PMID: 23041239 PMCID: PMC3490334
2067 PMID: 23709760 PMCID: PMC3756442
2070 22901061 PMC3533787
2083 PMID:23265842 PMC3569712
2124 23200744 PMC3552157
Name: PMID, Length: 194, dtype: object
def case2(id):
s = id.split()
if len(s) != 2:
return id
elif len(s[0]) == 8 and s[1][0:3]=="PMC":
# print s[0],s[1]
return s[0]
else:
return id
def isCase2(id):
s = id.split()
if len(s) != 2:
return False
elif len(s[0]) == 8 and s[1][0:3]=="PMC":
return True
else:
return False
df["isCase2"]=df.PMID.apply(isCase2)
pmcids = df[df.isCase2]["PMID"].apply(lambda x : x.split()[1])
df["PMID"] = df.PMID.apply(case2)
for i in pmcids.index:
df.at[i,"PMCID"] = pmcids[i]
df.drop("isCase2",axis=1,inplace=True)
df.PMID[df.PMID.str.contains("PMC")]
5 : PMC3805332
13 \n PMC3498934
14 PMID:22993091 PMC3447403
24 21624095 PMCID: PMC3734623
93 PMID:22961729 PMC3547296
94 PMID:22730171 PMC3556687
95 PMID:21472932 PMC3555362
107 PMID:23089748 PMC3535899
110 PMID:23817378 PMCID: PMC3754341
147 PMID:22326920 PMC3480643
164 PMCID:\n PMC3554041\n
165 PMC3413243
211 23931322 PMCID: PMC3736666
338 23121507 PMCID: PMC3596649
422 PMID22345357 PMC3539454
425 PMID:23483642 PMC3625746
426 23418011 PMCID: PMC3792637
431 22996943 PMCID: PMC3569615
434 23495205685.88 PMC3743159
468 PMID:23143153 PMC3566545
562 PMC3791366
596 PMID:22928509 PMC3707360
632 Monograph chapter, not in PMC
633 Monograph chapter, not in PMC
642 PMID:23220237 PMC3630327
758 PMID:22419580 PMC3487588
784 (PMCID:PMC3757918)
793 PMID23396536 PMC3652415
830 PMID:22988015 PMC3497410
834 PMID:23115042 PMC3536122
...
1502 PMID:23154538 PMC3717798
1537 PMC 3679597
1591 24039607PMC3764205
1612 23326615 PMCID: PMC3542187
1672 : PMC3487758
1687 24124519PMC3790821
1717 PMID: 23940704 PMCID: PMC3733773
1718 PMID: 23894442 PMCID: PMC3716603
1756 PMCID: PMC3491040
1762 PMID23251490 PMC3519623
1766 PMID:23613774 PMCID: PMC3632543
1770 23840430 : PMC3686795
1785 PMID: 24023766 PMCID: PMC3762823
1793 \n PMC3576415
1825 PMID: 23468629 PMCID: PMC3585132
1841 \nPMC3596319
1856 PMID: 23375993 PMCID: PMC3712186
1863 PMCID: PMC3465389
1892 PMID:23391734 PMC3581986
1907 PMID:22997008 PMC3569869
1935 22777780 PMCID: PMC3568905
1950 23422316 PMCID: PMC3710354
1993 PMID23287458 PMC3605587
2010 PMID:23623732 PMC3791409
2033 PMID: 23371065 PMCID: PMC3633812
2051 23576705, PMC3727040
2052 23804098 PMCID: PMC3724994
2062 PMID: 23041239 PMCID: PMC3490334
2067 PMID: 23709760 PMCID: PMC3756442
2083 PMID:23265842 PMC3569712
Name: PMID, Length: 103, dtype: object
Another pattern seems to be
PMID:XXXXXXXX PMCYYYYYYY
def case3(id):
s = id.split()
if len(s) != 2:
return id
elif (len(s[0]) == 12 or len(s[0]) == 13) and s[1][0:3]=="PMC":
# print s[0],s[1]
return s[0][5:]
else:
return id
def isCase3(id):
s = id.split()
if len(s) != 2:
return False
elif len(s[0]) == 13 and s[1][0:3]=="PMC":
return True
else:
return False
df["isCase3"] = df.PMID.apply(isCase3)
pmcids = df[df.isCase3]["PMID"].apply(lambda x : x.split()[1])
pmcids.at[1068] = pmcids.at[1068].replace("ID","")
df["PMID"] = df.PMID.apply(case3)
for i in pmcids.index:
df.at[i,"PMCID"] = pmcids[i]
df.drop("isCase3",axis=1,inplace=True)
df.PMID[df.PMID.str.contains("PMC")]
5 : PMC3805332
13 \n PMC3498934
24 21624095 PMCID: PMC3734623
110 PMID:23817378 PMCID: PMC3754341
164 PMCID:\n PMC3554041\n
165 PMC3413243
211 23931322 PMCID: PMC3736666
338 23121507 PMCID: PMC3596649
426 23418011 PMCID: PMC3792637
431 22996943 PMCID: PMC3569615
434 23495205685.88 PMC3743159
562 PMC3791366
632 Monograph chapter, not in PMC
633 Monograph chapter, not in PMC
784 (PMCID:PMC3757918)
868 PMID: 23209475 PMCID: PMC3504370
872 PMID:23570314 - PMC3634610
884 23319734 : PMC3672848
922 PMID: 23788479 PMCID: PMC3797641
958 : PMC3711325
960 PMC3516716
972 2339840 PMC3591641
986 PMCID: PMC3750171
990 23704797 PMCID: PMC3662417
1016 PMID: 23846817 PMCID: PMC3784648
1017 PMID: 23783094 : PMC3813945
1050 23324467 PMCID: PMC3709587
1062 23455506 PMCID: PMC3607399
1063 PMID:23475217 - PMC3619528
1064 PMID:23046967 PMCID: PMC3540259
...
1273 PMID:23518266 -PMC3839404
1287 PMID:23769710 PMCID: PMC3820028
1309 23256604 PMCID: PMC3587388
1456 PMID:23500098 -PMC3661980
1478 23595147 PMCID: PMC3675483
1479 PMID: 23771140 PMCID: PMC3753647
1491 PMID: 21948792 PMCID: PMC3245117
1537 PMC 3679597
1591 24039607PMC3764205
1612 23326615 PMCID: PMC3542187
1672 : PMC3487758
1687 24124519PMC3790821
1717 PMID: 23940704 PMCID: PMC3733773
1718 PMID: 23894442 PMCID: PMC3716603
1756 PMCID: PMC3491040
1766 PMID:23613774 PMCID: PMC3632543
1770 23840430 : PMC3686795
1785 PMID: 24023766 PMCID: PMC3762823
1793 \n PMC3576415
1825 PMID: 23468629 PMCID: PMC3585132
1841 \nPMC3596319
1856 PMID: 23375993 PMCID: PMC3712186
1863 PMCID: PMC3465389
1935 22777780 PMCID: PMC3568905
1950 23422316 PMCID: PMC3710354
2033 PMID: 23371065 PMCID: PMC3633812
2051 23576705, PMC3727040
2052 23804098 PMCID: PMC3724994
2062 PMID: 23041239 PMCID: PMC3490334
2067 PMID: 23709760 PMCID: PMC3756442
Name: PMID, Length: 67, dtype: object
def case4(id):
s = id.split()
if len(s) != 4:
return id
elif s[0]=="PMID:" and s[2]=="PMCID:":
# print s[0],s[1]
return s[1]
else:
return id
def isCase4(id):
s = id.split()
if len(s) != 4:
return False
elif s[0]=="PMID:" and s[2]=="PMCID:":
return True
else:
return False
df["isCase4"] = df.PMID.apply(isCase4)
pmcids = df[df.isCase4]["PMID"].apply(lambda x : x.split()[-1])
df["PMID"] = df.PMID.apply(case4)
for i in pmcids.index:
df.at[i,"PMCID"] = pmcids[i]
df.drop("isCase4",axis=1,inplace=True)
df.PMID[df.PMID.str.contains("PMC")]
5 : PMC3805332
13 \n PMC3498934
24 21624095 PMCID: PMC3734623
110 PMID:23817378 PMCID: PMC3754341
164 PMCID:\n PMC3554041\n
165 PMC3413243
211 23931322 PMCID: PMC3736666
338 23121507 PMCID: PMC3596649
426 23418011 PMCID: PMC3792637
431 22996943 PMCID: PMC3569615
434 23495205685.88 PMC3743159
562 PMC3791366
632 Monograph chapter, not in PMC
633 Monograph chapter, not in PMC
784 (PMCID:PMC3757918)
872 PMID:23570314 - PMC3634610
884 23319734 : PMC3672848
958 : PMC3711325
960 PMC3516716
972 2339840 PMC3591641
986 PMCID: PMC3750171
990 23704797 PMCID: PMC3662417
1017 PMID: 23783094 : PMC3813945
1050 23324467 PMCID: PMC3709587
1062 23455506 PMCID: PMC3607399
1063 PMID:23475217 - PMC3619528
1064 PMID:23046967 PMCID: PMC3540259
1098 23318955 PMCID: PMC3593212
1105 23474851 : PMC3664183
1132 23447590 : PMC3601669
1211 22875964 PMCID: PMC3457148
1218 23567253 : PMC3641668
1243 \nPMC3691584
1273 PMID:23518266 -PMC3839404
1287 PMID:23769710 PMCID: PMC3820028
1309 23256604 PMCID: PMC3587388
1456 PMID:23500098 -PMC3661980
1478 23595147 PMCID: PMC3675483
1537 PMC 3679597
1591 24039607PMC3764205
1612 23326615 PMCID: PMC3542187
1672 : PMC3487758
1687 24124519PMC3790821
1756 PMCID: PMC3491040
1766 PMID:23613774 PMCID: PMC3632543
1770 23840430 : PMC3686795
1793 \n PMC3576415
1841 \nPMC3596319
1863 PMCID: PMC3465389
1935 22777780 PMCID: PMC3568905
1950 23422316 PMCID: PMC3710354
2051 23576705, PMC3727040
2052 23804098 PMCID: PMC3724994
Name: PMID, dtype: object
def case5(id):
s = id.split()
if len(s) != 3:
return id
elif s[0][0:5]=="PMID:" and s[1]=="PMCID:":
# print s[0],s[1]
return s[0][5:]
else:
return id
def isCase5(id):
s = id.split()
if len(s) != 3:
return False
elif len(s[0]) == 13 and s[1][0:3]=="PMC":
return True
else:
return False
df["isCase5"] = df.PMID.apply(isCase5)
pmcids = df[df.isCase5]["PMID"].apply(lambda x : x.split()[-1])
df["PMID"] = df.PMID.apply(case5)
for i in pmcids.index:
df.at[i,"PMCID"] = pmcids[i]
df.drop("isCase5",axis=1,inplace=True)
df.PMID[df.PMID.str.contains("PMC")]
5 : PMC3805332
13 \n PMC3498934
24 21624095 PMCID: PMC3734623
164 PMCID:\n PMC3554041\n
165 PMC3413243
211 23931322 PMCID: PMC3736666
338 23121507 PMCID: PMC3596649
426 23418011 PMCID: PMC3792637
431 22996943 PMCID: PMC3569615
434 23495205685.88 PMC3743159
562 PMC3791366
632 Monograph chapter, not in PMC
633 Monograph chapter, not in PMC
784 (PMCID:PMC3757918)
872 PMID:23570314 - PMC3634610
884 23319734 : PMC3672848
958 : PMC3711325
960 PMC3516716
972 2339840 PMC3591641
986 PMCID: PMC3750171
990 23704797 PMCID: PMC3662417
1017 PMID: 23783094 : PMC3813945
1050 23324467 PMCID: PMC3709587
1062 23455506 PMCID: PMC3607399
1063 PMID:23475217 - PMC3619528
1098 23318955 PMCID: PMC3593212
1105 23474851 : PMC3664183
1132 23447590 : PMC3601669
1211 22875964 PMCID: PMC3457148
1218 23567253 : PMC3641668
1243 \nPMC3691584
1273 PMID:23518266 -PMC3839404
1309 23256604 PMCID: PMC3587388
1456 PMID:23500098 -PMC3661980
1478 23595147 PMCID: PMC3675483
1537 PMC 3679597
1591 24039607PMC3764205
1612 23326615 PMCID: PMC3542187
1672 : PMC3487758
1687 24124519PMC3790821
1756 PMCID: PMC3491040
1770 23840430 : PMC3686795
1793 \n PMC3576415
1841 \nPMC3596319
1863 PMCID: PMC3465389
1935 22777780 PMCID: PMC3568905
1950 23422316 PMCID: PMC3710354
2051 23576705, PMC3727040
2052 23804098 PMCID: PMC3724994
Name: PMID, dtype: object
def case6(id):
s = id.split()
if len(s) != 3:
return id
elif len(s[0])==8 and s[1]=="PMCID:":
# print s[0],s[1]
return s[0]
else:
return id
def isCase6(id):
s = id.split()
if len(s) != 3:
return False
elif len(s[0])==8 and s[1]=="PMCID:":
# print s[0],s[1]
return True
else:
return False
df["isCase6"] = df.PMID.apply(isCase6)
pmcids = df[df.isCase6]["PMID"].apply(lambda x : x.split()[-1])
df["PMID"] = df.PMID.apply(case6)
for i in pmcids.index:
df.at[i,"PMCID"] = pmcids[i]
df.drop("isCase6",axis=1,inplace=True)
df.PMID[df.PMID.str.contains("PMC")]
5 : PMC3805332
13 \n PMC3498934
164 PMCID:\n PMC3554041\n
165 PMC3413243
434 23495205685.88 PMC3743159
562 PMC3791366
632 Monograph chapter, not in PMC
633 Monograph chapter, not in PMC
784 (PMCID:PMC3757918)
872 PMID:23570314 - PMC3634610
884 23319734 : PMC3672848
958 : PMC3711325
960 PMC3516716
972 2339840 PMC3591641
986 PMCID: PMC3750171
1017 PMID: 23783094 : PMC3813945
1063 PMID:23475217 - PMC3619528
1105 23474851 : PMC3664183
1132 23447590 : PMC3601669
1218 23567253 : PMC3641668
1243 \nPMC3691584
1273 PMID:23518266 -PMC3839404
1456 PMID:23500098 -PMC3661980
1537 PMC 3679597
1591 24039607PMC3764205
1672 : PMC3487758
1687 24124519PMC3790821
1756 PMCID: PMC3491040
1770 23840430 : PMC3686795
1793 \n PMC3576415
1841 \nPMC3596319
1863 PMCID: PMC3465389
2051 23576705, PMC3727040
Name: PMID, dtype: object
Just do the rest by hand
df.at[5,"PMID"] = "Unknown"
df.at[5,"PMCID"] = "PMC3805332"
df.at[13,"PMID"] = "Unknown"
df.at[13,"PMCID"] = "PMC3498934"
df.at[164,"PMID"] = "Unknown"
df.at[164,"PMCID"] = "PMC3554041"
df.at[165,"PMID"] = "Unknown"
df.at[165,"PMCID"] = "PMC3413243"
df.at[434,"PMID"] = "23495205" # looked in pubmed
df.at[434,"PMCID"] = "PMC3743159"
df.at[562,"PMID"] = "Unknown"
df.at[562,"PMCID"] = "PMC3791366"
df.at[632,"PMID"] = "Unknown"
df.at[632,"PMCID"] = "Unknown"
df.at[633,"PMID"] = "Unknown"
df.at[633,"PMCID"] = "Unknown"
df.at[784,"PMID"] = "Unknown"
df.at[784,"PMCID"] = "PMC3757918"
df.at[872,"PMID"] = "23570314"
df.at[872,"PMCID"] = "PMC3634610"
df.at[884,"PMID"] = "23319734"
df.at[884,"PMCID"] = "PMC3672848"
df.at[958,"PMID"] = "Unknown"
df.at[958,"PMCID"] = "PMC3711325"
df.at[960,"PMID"] = "Unknown"
df.at[960,"PMCID"] = "PMC3516716"
df.at[972,"PMID"] = "23329840" # looked in pubmed, the 2 was missing!
df.at[972,"PMCID"] = "PMC3591641"
df.at[986,"PMID"] = "Unknown"
df.at[986,"PMCID"] = "PMC3750171"
df.at[1017,"PMID"] = "23783094"
df.at[1017,"PMCID"] = "PMC3813945"
df.at[1063,"PMID"] = "23475217"
df.at[1063,"PMCID"] = "PMC3619528"
df.at[1105,"PMID"] = "23474851"
df.at[1105,"PMCID"] = "PMC3664183"
df.at[1132,"PMID"] = "23447590"
df.at[1132,"PMCID"] = "PMC3601669"
df.at[1218,"PMID"] = "23567253"
df.at[1218,"PMCID"] = "PMC3641668"
df.at[1243,"PMID"] = "Unknown"
df.at[1243,"PMCID"] = "PMC3691584"
df.at[1273,"PMID"] = "23518266"
df.at[1273,"PMCID"] = "PMC3839404"
df.at[1456,"PMID"] = "23500098"
df.at[1456,"PMCID"] = "PMC3661980"
df.at[1537,"PMID"] = "Unknown"
df.at[1537,"PMCID"] = "PMC3679597"
df.at[1591,"PMID"] = "24039607"
df.at[1591,"PMCID"] = "PMC3764205"
df.at[1672,"PMID"] = "Unknown"
df.at[1672,"PMCID"] = "PMC3487758"
df.at[1687,"PMID"] = "24124519"
df.at[1687,"PMCID"] = "PMC3790821"
df.at[1756,"PMID"] = "Unknown"
df.at[1756,"PMCID"] = "PMC3491040"
df.at[1770,"PMID"] = "23840430"
df.at[1770,"PMCID"] = "PMC3686795"
df.at[1793,"PMID"] = "Unknown"
df.at[1793,"PMCID"] = "PMC3576415"
df.at[1841,"PMID"] = "Unknown"
df.at[1841,"PMCID"] = "PMC3596319"
df.at[1863,"PMID"] = "Unknown"
df.at[1863,"PMCID"] = "PMC3465389"
df.at[2051,"PMID"] = "23576705"
df.at[2051,"PMCID"] = "PMC3727040"
df.PMID[df.PMID.str.contains(":")]
17 PMID: 23828613 (July 2013 Epub)
77 PMID: 23409903 23409903
78 PMID:23331923
114 PMID:23706023
125 PMID:23971731
166 PMID: 23459248
187 PMID: 23245760
188 PMID: 23452663
189 PMID: 23510580
234 PMID:23442822
327 PMID: 23822584
334 PMID: 23927070
349 PMID:23137731
367 PMID: 23966160
369 PMID:23974100
387 PMID: 23994477
394 PMID: 23041313 23041313
413 PMID:24047602
424 PMID:24009110
432 PMID: 23918316
495 PMID: 23267662
499 PMID: 23935057
501 PMID:24118397
513 PMID: 23861059
552 PMID: 22171404
648 PMID: 23879611
654 PMID: 23118027
670 PMID : 23763300
716 PMID: 23256812
717 PMID: 24024949
...
1229 PMID:23545320
1272 PMID: 23494299
1306 PMID:23182425
1307 PMID:23200636
1312 PMID:24125554
1314 PMID: 23980694
1315 PMID: 23750903
1316 PMID: 23888912
1374 PMID:23639206
1381 PMID: 18755526
1442 PMID: 23295962
1446 PMID:24056298
1475 PMID:24038356
1498 PMID: 23686316
1499 PMID: 23708655
1500 PMID:23975431
1527 PMID: 23931634 Aug 2013 Epub date
1541 PMID:23023652
1543 PMID:23898885
1678 PMID:23300533
1816 PMID:23633946
1857 PMID: 22964003
1883 PMID: 23650371 23650371
1948 PMID:23635806
1953 PMID: 23236081
2001 PMID: 23703895
2018 PMID: 23649934
2063 PMID : 23773811
2075 PMID: 23305527
2109 PMID: 24035434
Name: PMID, Length: 85, dtype: object
def removeColon(id):
if len(id)==13 and id[0:5]=="PMID:":
return id[6:]
else:
return id
df["PMID"] = df["PMID"].apply(removeColon)
def removeColonSpace(id):
if len(id)==14 and id[0:6]=="PMID: ":
return id[7:]
else:
return id
df["PMID"] = df["PMID"].apply(removeColonSpace)
df.PMID[df.PMID.str.contains(":")]
17 PMID: 23828613 (July 2013 Epub)
77 PMID: 23409903 23409903
166 PMID: 23459248
394 PMID: 23041313 23041313
654 PMID: 23118027
670 PMID : 23763300
741 PMID: 23703681 (May 2013 Epub)
907 PMID: 23846567 (Epub July 2013)
1036 PMID: 22360292
1067 PMID: 23945372
1069 PMID:24048963 (Epub Sept 2013)
1076 PMID: 23962810 Aug 2013 Epub
1527 PMID: 23931634 Aug 2013 Epub date
1883 PMID: 23650371 23650371
2063 PMID : 23773811
Name: PMID, dtype: object
df.at[17,"PMID"] = "23828613"
df.at[77,"PMID"] = "23409903"
df.at[166,"PMID"] = "23459248"
df.at[394,"PMID"] = "23041313"
df.at[654,"PMID"] = "23118027"
df.at[670,"PMID"] = "23763300"
df.at[741,"PMID"] = "23703681"
df.at[907,"PMID"] = "23846567"
df.at[1036,"PMID"] = "22360292"
df.at[1067,"PMID"] = "23945372"
df.at[1069,"PMID"] = "24048963"
df.at[1076,"PMID"] = "23962810"
df.at[1527,"PMID"] = "23931634"
df.at[1883,"PMID"] = "23650371"
df.at[2063,"PMID"] = "23773811"
df.PMID[ (df.PMID.str.len()!=8) & (df.PMID != "Unknown") ]
22 3535376
25 In Process
35 print published August 2013
37 3748854
39 3633780
40 In Process
52 3516067
61 3813311
69 3567269
71 published Sept 2013
72 3947303
75 3708126
78 3331923
114 3706023
116 Not yet available
125 3971731
127 In Process
148 3685266
172 3654172
176 3847282
187 3245760
188 3452663
189 3510580
198 3558749
201 3744075
234 3442822
237 3526451
238 3681581
239 3708772
244 3639180
...
1911 e-pub
1912 in press - due August 13
1929 543219
1933 PMID23824939
1934 Not yet available
1944 3281495
1945 3651613
1948 3635806
1953 3236081
1958 -
1974 Epub ahead of print - April 2013
1975 Epub ahead of print pub Dec 2012, print in pre...
1988 3778404
1993 3287458
1995 PMID23278317
1997 Not yet available
2000 22897899 MC3489778
2001 3703895
2003 3223522
2004 3813980
2016 3635120
2017 -
2018 3649934
2037 Not yet available
2073 3744763
2075 3305527
2079 pub Aug 2013
2086 print in press
2109 4035434
2123 pub Aug 2013
Name: PMID, Length: 350, dtype: object
df.PMID[ (df.PMID.str.len()!=8) & (df.PMID != "Unknown") & (df.PMID.str.startswith("PMID"))]
1263 PMID24069883
1264 PMID24069886
1265 PMID24069913
1521 PMID23790101
1707 PMID22970197
1906 PMID23184293
1933 PMID23824939
1995 PMID23278317
Name: PMID, dtype: object
def stripPMID(id):
if len(id)==12 and id[0:4]=="PMID":
return id[5:]
else:
return id
df["PMID"] = df["PMID"].apply(stripPMID)
df.PMID[ (df.PMID.str.len()!=8) & (df.PMID != "Unknown")]
22 3535376
25 In Process
35 print published August 2013
37 3748854
39 3633780
40 In Process
52 3516067
61 3813311
69 3567269
71 published Sept 2013
72 3947303
75 3708126
78 3331923
114 3706023
116 Not yet available
125 3971731
127 In Process
148 3685266
172 3654172
176 3847282
187 3245760
188 3452663
189 3510580
198 3558749
201 3744075
234 3442822
237 3526451
238 3681581
239 3708772
244 3639180
...
1911 e-pub
1912 in press - due August 13
1929 543219
1933 3824939
1934 Not yet available
1944 3281495
1945 3651613
1948 3635806
1953 3236081
1958 -
1974 Epub ahead of print - April 2013
1975 Epub ahead of print pub Dec 2012, print in pre...
1988 3778404
1993 3287458
1995 3278317
1997 Not yet available
2000 22897899 MC3489778
2001 3703895
2003 3223522
2004 3813980
2016 3635120
2017 -
2018 3649934
2037 Not yet available
2073 3744763
2075 3305527
2079 pub Aug 2013
2086 print in press
2109 4035434
2123 pub Aug 2013
Name: PMID, Length: 350, dtype: object
df.at[2000,"PMID"]=22897899
df.at[2000,"PMCID"]="PMC3489778"
OK, I think that’s all we can recover for the ids without requerying the database.
df.head()
| PMCID | Publisher | Journal Title | Article Title | TotalCost | PMID | |
|---|---|---|---|---|---|---|
| 0 | PMC3378987 | Elsevier | Academy of Nutrition and Dietetics | Parent support and parent mediated behaviours ... | 2379.54 | Unknown |
| 1 | PMC3780468 | ACS (Amercian Chemical Society) Publications | ACS Chemical Biology | A Novel Allosteric Inhibitor of the Uridine Di... | 1294.59 | Unknown |
| 2 | PMC3621575 | ACS (Amercian Chemical Society) Publications | ACS Chemical Biology | Chemical proteomic analysis reveals the drugab... | 1294.78 | Unknown |
| 3 | Unknown | American Chemical Society | ACS Chemical Biology | Discovery of β2 Adrenergic Receptor Ligands Us... | 947.07 | Unknown |
| 4 | PMC3833349 | American Chemical Society | ACS Chemical Biology | Discovery of an allosteric inhibitor binding s... | 1267.76 | 24015914 |
df.Publisher[df.Publisher.str.contains("sevier")] = "Elsevier"
/Users/skaplan/Library/Python/2.7/lib/python/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
"""Entry point for launching an IPython kernel.
df.Publisher[df.Publisher.str.contains("iley")].value_counts()
Wiley 136
Wiley-Blackwell 56
Wiley Subscription Services Inc. 13
Wiley 12
Wiley Subscription Services 8
John Wiley & Sons Ltd 8
John Wiley & Sons 6
Wiley-VCH 6
John Wiley 4
Wiley Blackwell 4
John Wiley & Sons Inc 2
Wiley Subscription Services Inc 2
John Wiley and Sons Ltd 1
Blackwell Publishing Ltd/Wiley 1
Wiley VCH 1
Wiley Subscription Serviices Inc 1
John Wiley & Sons, Inc. 1
Wiley-Blackwell, John Wiley & Sons 1
Wiley/Blackwell 1
Wiley & Son 1
Wiley Online Library 1
John Wiley and Sons 1
Name: Publisher, dtype: int64
df.Publisher[df.Publisher.str.contains("iley")] = "Wiley-Blackwell"
/Users/skaplan/Library/Python/2.7/lib/python/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
"""Entry point for launching an IPython kernel.
df.Publisher.value_counts()
Elsevier 403
Public Library of Science 278
Wiley-Blackwell 267
Springer 81
Oxford University Press 77
OUP 56
ASBMB 46
Nature Publishing Group 45
BioMed Central 40
BMC 26
Nature 24
Frontiers 23
BMJ 23
Royal Society 22
Cambridge University Press 20
Company of Biologists 18
American Society for Biochemistry and Molecular Biology 18
Dartmouth Journal Services 17
Oxford Journals 16
American Society for Microbiology 16
PLoS 15
National Academy of Sciences 15
American Chemical Society 14
Society for General Microbiology 14
Landes Bioscience 14
BMJ Group 13
American Psychological Association 13
Sage 12
Taylor & Francis 11
Society for Neuroscience 11
...
The American Physiological Society 1
American Psychiatric Association 1
Oxford Univesity Press 1
American Speech-Language-Hearing Association 1
ASBMC /CENVEO 1
Informa Healthcare communications 1
Wliey-Blackwell 1
Benthan Science Publishers 1
Company of Biologists Ltd 1
The American Society for Biochemistry and Molecular Biology, Inc 1
SOCIETY OF NEURO SCIENCES 1
Cold Spring Harbor 1
International Union Against tuberculosis and Lung Disease 1
Association for Research in Vision & Ophthalmology 1
Springer-Verlag GmbH, Heidelberger Platz 3, D-14197 Berlin 1
J Med Internet Research 1
American Chemical Society Publications 1
The company of Biologists 1
public.service.co.uk 1
American Society for Microbiology 1
Sage Publications Inc 1
BMJ Publishing Group Ltd & British Thoracic Society 1
Springer 1
IOS Press 1
Impact Journals LLC 1
Sage Publications 1
Wolters Kluwers 1
ASM (American Society for Microbiology) 1
CADMUS JOURNAL SERVICES 1
Hindawi 1
Name: Publisher, Length: 271, dtype: int64
df.Publisher[df.Publisher.str.contains("pringer")].value_counts()
Springer 81
Springer - Verlag GMBH 2
Springer-Verlag GmbH 2
Springer-Verlag GmbH, Heidelberger Platz 3, D-14197 Berlin 1
Springer Verlag 1
Springer 1
Springer Science + Business Media 1
Springer-Veriag GmbH 1
Springer-Verlag GMBH & Ci 1
Humana Press (Springer Imprint) 1
Name: Publisher, dtype: int64
df.Publisher[df.Publisher.str.contains("pringer")]="Springer"
/Users/skaplan/Library/Python/2.7/lib/python/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
"""Entry point for launching an IPython kernel.
df.Publisher[df.Publisher.str.contains("xford")].value_counts()
Oxford University Press 77
Oxford Journals 16
Oxford University Press (OUP) 4
Oxford Univ Press 4
Oxford University Press\n 3
Oxford University Press 3
Oxford Univesity Press 1
Oxford Journals (OUP) 1
Name: Publisher, dtype: int64
df.Publisher[df.Publisher.str.contains("xford")]="Oxford University Press"
/Users/skaplan/Library/Python/2.7/lib/python/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
"""Entry point for launching an IPython kernel.
df.Publisher[df.Publisher=="OUP"]="Oxford University Press"
/Users/skaplan/Library/Python/2.7/lib/python/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
"""Entry point for launching an IPython kernel.
df.Publisher[df.Publisher=="ASBMB"]="American Society for Biochemistry and Molecular Biology"
/Users/skaplan/Library/Python/2.7/lib/python/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
"""Entry point for launching an IPython kernel.
df.Publisher[df.Publisher.str.contains("BMJ")] = "BMJ"
/Users/skaplan/Library/Python/2.7/lib/python/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
"""Entry point for launching an IPython kernel.
df.Publisher.value_counts()
Elsevier 403
Public Library of Science 278
Wiley-Blackwell 267
Oxford University Press 165
Springer 92
American Society for Biochemistry and Molecular Biology 64
BMJ 56
Nature Publishing Group 45
BioMed Central 40
BMC 26
Nature 24
Frontiers 23
Royal Society 22
Cambridge University Press 20
Company of Biologists 18
Dartmouth Journal Services 17
American Society for Microbiology 16
PLoS 15
National Academy of Sciences 15
American Chemical Society 14
Landes Bioscience 14
Society for General Microbiology 14
American Psychological Association 13
Sage 12
Taylor & Francis 11
Society for Neuroscience 11
Portland Press Ltd 10
Portland Press 10
BioMed Central 9
Biomed Central 8
...
Future Medicine Ltd 1
IOS Press 1
Darmouth Journal Services 1
The Boulevard 1
Journal of Visualized Experiments 1
Society for Neurosciences 1
The American Physiological Society 1
American Psychiatric Association 1
ASBMC /CENVEO 1
Impact Journals LLC 1
Transcript Verlag 1
The Journal of Visualized Experiments 1
SOCIETY OF NEURO SCIENCES 1
Palgrave MacMillan 1
American Soc for Biochemistry and Molecular Biology 1
Society of General Microbiology 1
AGA Institute 1
Royal Society for Chemistry 1
Cambridge Uni Press 1
Informa Healthcare 1
International Union Against tuberculosis and Lung Disease 1
Sage Publications Inc 1
Association for Research in Vision & Ophthalmology 1
Policy Press 1
THE COMPANY OF BIOLOGISTS 1
Federation of the American Society of Experimental Biology 1
American Chemical Society Publications 1
public.service.co.uk 1
American Public Health Association 1
American Society for Microbiology 1
Name: Publisher, Length: 244, dtype: int64
df.Publisher[(df.Publisher.str.contains("ACS")) | (df.Publisher.str.contains("Chemical"))] = "American Chemical Society"
/Users/skaplan/Library/Python/2.7/lib/python/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
"""Entry point for launching an IPython kernel.
df.Publisher.value_counts()
Elsevier 403
Public Library of Science 278
Wiley-Blackwell 267
Oxford University Press 165
Springer 92
American Society for Biochemistry and Molecular Biology 64
BMJ 56
Nature Publishing Group 45
BioMed Central 40
American Chemical Society 33
BMC 26
Nature 24
Frontiers 23
Royal Society 22
Cambridge University Press 20
Company of Biologists 18
Dartmouth Journal Services 17
American Society for Microbiology 16
PLoS 15
National Academy of Sciences 15
Society for General Microbiology 14
Landes Bioscience 14
American Psychological Association 13
Sage 12
Society for Neuroscience 11
Taylor & Francis 11
Portland Press Ltd 10
Portland Press 10
BioMed Central 9
CUP 8
...
Future Medicine Ltd 1
NATURE PUBLISHING GROUP LTD 1
The Boulevard 1
Journal of Visualized Experiments 1
Society for Neurosciences 1
The American Physiological Society 1
American Psychiatric Association 1
National Academy of Sciences 1
ASBMC /CENVEO 1
Informa Healthcare communications 1
Wliey-Blackwell 1
Cold Spring Harbor Publications 1
The royal Society 1
CADMUS JOURNAL SERVICES 1
public.service.co.uk 1
Federation of the American Society of Experimental Biology 1
Policy Press 1
THE COMPANY OF BIOLOGISTS 1
University of the Basque Country Press 1
Cadmus 1
American Public Health Association 1
American Society for Microbiology 1
Darmouth Journal Services 1
Sage Publications Inc 1
Transcript Verlag 1
IOS Press 1
Impact Journals LLC 1
Sage Publications 1
Wolters Kluwers 1
ASBMB/Cadmus 1
Name: Publisher, Length: 239, dtype: int64
df.Publisher[df.Publisher.str.contains("BMC")] = "BioMed Central"
/Users/skaplan/Library/Python/2.7/lib/python/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
"""Entry point for launching an IPython kernel.
df.head()
| PMCID | Publisher | Journal Title | Article Title | TotalCost | PMID | |
|---|---|---|---|---|---|---|
| 0 | PMC3378987 | Elsevier | Academy of Nutrition and Dietetics | Parent support and parent mediated behaviours ... | 2379.54 | Unknown |
| 1 | PMC3780468 | American Chemical Society | ACS Chemical Biology | A Novel Allosteric Inhibitor of the Uridine Di... | 1294.59 | Unknown |
| 2 | PMC3621575 | American Chemical Society | ACS Chemical Biology | Chemical proteomic analysis reveals the drugab... | 1294.78 | Unknown |
| 3 | Unknown | American Chemical Society | ACS Chemical Biology | Discovery of β2 Adrenergic Receptor Ligands Us... | 947.07 | Unknown |
| 4 | PMC3833349 | American Chemical Society | ACS Chemical Biology | Discovery of an allosteric inhibitor binding s... | 1267.76 | 24015914 |
df["Journal"] = df["Journal Title"]
df.drop("Journal Title",axis=1,inplace=True)
df.head()
| PMCID | Publisher | Article Title | TotalCost | PMID | Journal | |
|---|---|---|---|---|---|---|
| 0 | PMC3378987 | Elsevier | Parent support and parent mediated behaviours ... | 2379.54 | Unknown | Academy of Nutrition and Dietetics |
| 1 | PMC3780468 | American Chemical Society | A Novel Allosteric Inhibitor of the Uridine Di... | 1294.59 | Unknown | ACS Chemical Biology |
| 2 | PMC3621575 | American Chemical Society | Chemical proteomic analysis reveals the drugab... | 1294.78 | Unknown | ACS Chemical Biology |
| 3 | Unknown | American Chemical Society | Discovery of β2 Adrenergic Receptor Ligands Us... | 947.07 | Unknown | ACS Chemical Biology |
| 4 | PMC3833349 | American Chemical Society | Discovery of an allosteric inhibitor binding s... | 1267.76 | 24015914 | ACS Chemical Biology |
df.Journal.value_counts()
PLoS One 92
PLoS ONE 62
Journal of Biological Chemistry 48
Nucleic Acids Research 21
Proceedings of the National Academy of Sciences 19
PLoS Neglected Tropical Diseases 18
Human Molecular Genetics 18
Nature Communications 17
PLoS Genetics 15
PLoS Pathogens 15
Neuroimage 15
PLOS ONE 14
BMC Public Health 14
Brain 14
NeuroImage 14
Movement Disorders 13
Biochemical Journal 12
Developmental Cell 12
Journal of Neuroscience 12
Journal of General Virology 11
BMJ 10
Current Biology 10
PLOS One 10
Cell Reports 9
Neuron 9
Journal of Physiology 8
Journal of Cell Science 8
European Journal of Immunology 8
Molecular Microbiology 8
Hepatology 8
..
Public Service Review 1
Frontiers in Neurorobotics 1
Frontiers in Decision Neuroscience 1
Cognitive Therapy and Research 1
PLoS 1
Current Opinion Microbiology 1
Int Journal for Parasitology 1
Frontiers in Invertebrate Physiology 1
Synapse 1
PNTD 1
Journal of Archaeological Science 1
Journal of Alzheimer Disease 1
International Immunology 1
The journal of Biological Chemistry 1
EvoDevo 1
Reproductive Sciences 1
International Reviews of Immunology 1
Theranostics 1
Journal of Epidemiology & Community Health 1
ASN Neuro 1
Clinical Radiology 1
Speech Language and Hearing Research 1
PLoS Medicine Journal 1
Cerebral Cortex print 1
Tissue Engineering: part A 1
Emotion 1
Arthritis Research and Therapy 1
The Vet. Journal 1
Journal of Transport Geography 1
American Jnl Epidemiology 1
Name: Journal, Length: 984, dtype: int64
df.Journal[(df.Journal.str.lower() == "plos") | (df.Journal.str.lower() == "plos 1") | (df.Journal.str.lower() == "plos one")] = "PLoS One"
/Users/skaplan/Library/Python/2.7/lib/python/site-packages/ipykernel_launcher.py:1: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
"""Entry point for launching an IPython kernel.
df.head()
| PMCID | Publisher | Article Title | TotalCost | PMID | Journal | |
|---|---|---|---|---|---|---|
| 0 | PMC3378987 | Elsevier | Parent support and parent mediated behaviours ... | 2379.54 | Unknown | Academy of Nutrition and Dietetics |
| 1 | PMC3780468 | American Chemical Society | A Novel Allosteric Inhibitor of the Uridine Di... | 1294.59 | Unknown | ACS Chemical Biology |
| 2 | PMC3621575 | American Chemical Society | Chemical proteomic analysis reveals the drugab... | 1294.78 | Unknown | ACS Chemical Biology |
| 3 | Unknown | American Chemical Society | Discovery of β2 Adrenergic Receptor Ligands Us... | 947.07 | Unknown | ACS Chemical Biology |
| 4 | PMC3833349 | American Chemical Society | Discovery of an allosteric inhibitor binding s... | 1267.76 | 24015914 | ACS Chemical Biology |
df = df[["PMID","PMCID","Article Title","Journal","Publisher","TotalCost"]]
df.head()
| PMID | PMCID | Article Title | Journal | Publisher | TotalCost | |
|---|---|---|---|---|---|---|
| 0 | Unknown | PMC3378987 | Parent support and parent mediated behaviours ... | Academy of Nutrition and Dietetics | Elsevier | 2379.54 |
| 1 | Unknown | PMC3780468 | A Novel Allosteric Inhibitor of the Uridine Di... | ACS Chemical Biology | American Chemical Society | 1294.59 |
| 2 | Unknown | PMC3621575 | Chemical proteomic analysis reveals the drugab... | ACS Chemical Biology | American Chemical Society | 1294.78 |
| 3 | Unknown | Unknown | Discovery of β2 Adrenergic Receptor Ligands Us... | ACS Chemical Biology | American Chemical Society | 947.07 |
| 4 | 24015914 | PMC3833349 | Discovery of an allosteric inhibitor binding s... | ACS Chemical Biology | American Chemical Society | 1267.76 |
df.TotalCost.hist(bins=30)
<matplotlib.axes._subplots.AxesSubplot at 0x110fe5350>
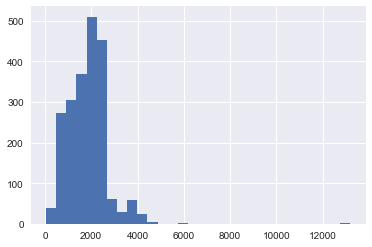
Whoa, what study cost more than 12000 GBP?!?
df[df.TotalCost>12000]
| PMID | PMCID | Article Title | Journal | Publisher | TotalCost | |
|---|---|---|---|---|---|---|
| 1341 | Unknown | Unknown | Fungal Disease in Britain and the United State... | NaN | MacMillan | 13200.0 |
df[df.TotalCost>12000]["Article Title"][1341]
u'Fungal Disease in Britain and the United States 1850-2000'